An interesting video about some challenges also DNA sequencing will have to answer.
Genetic Genealogy DTC DNA-Tests overview 2024
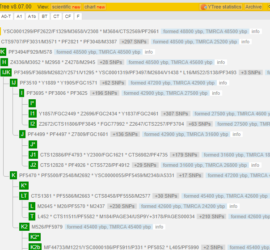
Update May 2024 Find relatives (autosomal & X-DNA) Statistically, every individual inherits at least one DNA segment from each of their great-great-grandparents (4th generation, 16 ancestors). Beginning with the 5th generation the genetic ancestors diminish: 8th generation 220/256 ancestors, 16th generation 1018/65536 ancestors (Source Bob Jenkins, see also Anzahl genetische Vorfahren…). This means that it […]