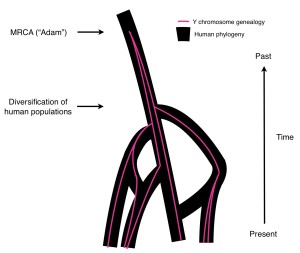
hypothetical Y-chromosome genealogy superimposed on a hypothetical human phylogeny (by Joe Pickrell, jkplab.org)
The topic when and where the most recent common ancestor (MRCA) of all human Y-chromosomes (somehow misleading but popularly called “Y-Adam”), inherited paternally from father to sons, did live, is one of the hottest topics in the population genetics field and affects also research and tools used by genetic genealogists focusing on the paternal lineage (Y-MRCA). One of the most important discoveries was made by Bonnie Schrack and genetic genealogists in Hg A: they found a very early lineage with a haplotype very different from everything known with origin in Cameroon. Low-res Y-sequencing of this paternal lineages eventually lead to the placing of this lineage now called A00 on the human Y-tree (see Mendez et al. 2013 and Skeleton Y-chr. phylogeny). We are still waiting for papers with new high-res sequencing data from A00 lineages. Bonnie Schrack is still active in this process and hopefully can deliver one of the first publications with her crowdfunding science-based research projects on A00 in Cameroon. The reason of this blog post is the publication of a critique by Elhaik, Klyosov et al. 2014 (without their own primary data) to Mendez et al. 2013 in a well-known journal (behind a paywall). See interview with co-author Dan Graur including a slideshare. Many things are confusing and of concern in this critique raised by authors without leadership in the Y-chromosome research. Let’s list some of them:
- the method to build the ‘own hypothesis’ (208 kya) on the data of the critizised paper and the ability to publish it peer-reviewed as ‘discovery’, even if Mendez et al. report themselves that using a higher mutation rate (1.0×10-9) results in a more ‘reasonable’ estimate of the Y-TMRCA of 209 kya.
- no willingness to provide or mention other alternative models, views, calibrations, discussions and papers.
- an unfriendly way to represent the criticized paper results (and therefore his authors) as deliberatedly falsified or amateurish, to make the own hypothesis seem more important (see YouTube video by Elhaik). This procedure also distracts from the open and necessary scientific discussion of the paper results.
- instead of using meaningful words like “perhaps”, “likely”, “assuming” and synonyms for the own counter-hypothesis, focusing on the critique with words like “extraordinarily early”, “astonishing”, “space-time-paradox”, “Mendez et al. misled thee”, “cheating”, “invented a new branch of statistics”, “assumptions”, “data manipulation”, etc.
- activities and beliefs of the criticizing authors who differ fundamentally from the majority expert opinion and the need of the now published “anti-hypothesis” to strengthen their own “worldview”.
Back to topic – only by providing this data IMO we will have a much clearer picture of the age and mutation calibration of the human Y-phylogeny and true discoveries:
- Highest-Res Y-Sequences of all existing major lineages, especially A00-L1086, A0-V148, A1-M31, A2’3-L419 and B-M60 with proven genealogies over many generations of distance.
- High-Res Y-Sequences of many precisely dated ancient Y-DNA back to Neanderthals and Denisovans.
- Comparison of 1. and 2. and focus on the probability of correspondence of the genetic and archaeological age of clear expansion points, like the main Asio-Euro expansion of “anatomically modern humans (AMH)“, which is usually set in Y-phylogeny, with the Haplogroup CDEF-M168 as the progenitor.
For the necessary discussion on the Y-mutation rate and age estimates there is a more topic focused and IMO correctly formulated and interesting critique by Wilson-Sayres 2013 (arXiv preprint open access). The comment is based on comparing estimates of male mutation bias using different chromosome comparisons (X/Autosome, Y/Autosome, X/Y) and observed diversity on the entire Y chromosome (1/10 of what is expected due to the effects of selection acting to reduce diversity). Mendez et al. uses incautiously autosomal substitution rates and 30 years per generation. While those critique points are all valid, Wilson-Sayres is assuming based on neutral population genetic theory (variability within species are not caused by natural selection but by random drift), that the TMRCA for the Y should be in line with TMRCA estimates for the mtDNA, 1/3 of the X and 1/4 of the autosomes. As I understand it, this is only correct if we always had only one population of DNA-ancestors of all modern living humans (no detectable hybridization since the split from the other great apes). See also from April 2014 Dienekes Blog Post Criticism of Y-chromosome Adam old age and Haldane’s Sieve post about Wilson-Sayres preprint with comments from W-S.
Collegue Vince Tilroe points to a very important aspect: AMH is based on existing ancient fossil evidence, so declaring an existing haplogroup as pre-human or non-human is very risky, as the term “Human” is philosophically derived and like AMH has nothing to do directly with genetic evidence.
If you are further interested in the topic of “Y-TMRCA compared to AMH-TMRCA” I suggest to read the blog post of Joe Pickrell @ jkplab.org debunking Elhaik et al. claims of “Our estimate excludes the possibility of introgression with most ancient hominine species.” as unreasonable and correctly saying that they “have confused themselves into thinking that [Y-Adam) must have been human.”. To this topic I would add Elhaik et al. do strongly refuse any form of hybridization close to the time when the “AMH” were formed, see the funny ‘biblical’ claim “Adam has never engaged in zoophilic relationships with other species.”. Aside from the unscientific appropriation of Adam this is simply illogical, since autosomal genetics already clearly shows that AMH much later had sexual contact with distinguishable species, such as the Neanderthals or Denisovans. Probably we will see critiquing publications on this evidence from the same authors. I’m awaiting more independent publications commenting Elhaik et al. At least a little entertainment in this humourless profession…
Update: reviews regarding Elhaik’s publications and his “scientific feuds”
- Razib Khan, Gene Expression, August 2012: Ashkenazi Jews are probably not descended from the Khazars
- Jon Entine, Forbes, May 2013: Israeli Researcher Challenges Jewish DNA links to Israel, Calls Those Who Disagree ‘Nazi Sympathizers’
- Rita Rubin, Forward, May 2013: ‘Jews a Race’ Genetic Theory Comes Under Fierce Attack by DNA Expert
