Recently Wim Penninx posted in the ISOGG Facebook group about his analysis of the known Y-DNA ancestors of Ashkenazi Jews (from where did they arrive in the area of the kingdom of Poland and the Polish-Lithuanian Commonwealth). The first quick look revealed a pearl of citizen science using a large collection of Direct to Consumer (DTC) Y-DNA data. As Wim encourages review and feedback I decided to have a better look and write down my taughts what I would change or do differently before doing a focused analysis on J2 Jewish lineages.
I had no time to look at the extensive data in full detail, so some of my comments and conclusions might be superficial or to revise with an update, maybe after Wim clears something out.
Update: Wim Pennix’s reply (28 Sep. 2014, thank you for allowing me to insert it here), my reply (30 Sep. 2014) and some minor edits added:
-
now established almost 70 groups … we will have a few more branches (10-20%)
Looks impressive. Comparison with the quantity found in publications would be interesting. I would omit the 40 in the study title.
-
I wanted a url that is stable and easy to remember. “TheAshkenazimProgenitors” ? I could also change the number. There is an uncertainty of of the amount of small branches, for which i have only one person now. I could also keep the url, but change the title.
For the URL maybe “AskenaziY” is interesting and short?
-
-
about 2000 Ashkenazim in this dataset
Very convenient that this ethnic group is among the most tested by FTDNA and with more Y-STR markers (67, also 111 available for some) and more Y-SNPs (sanger sequencing, chip testing and even NG-Y-sequencing) then commonly used testing in forensic studies. No better data available elsewhere probably.
-
🙂
-
-
I try to determine the percentages of the branches in the following groups:
- Groups with people that probably originated in the area of Judea and spread to Spain (Sephardic) and later moved to the Polish-Lithuanian area.
- People who converted to the Jewish religion in the area of the Polish-Lithuanian area.
- Possible origin of the Khazars
Instead of Judea later the more consistent origin “Middle East” is used. I think that should be corrected also here, if there are no subgroups clearly recognizable as Judean or Levantic (without ancient DNA probably very speculative). If Sephardic is strictly connected to detectable presence in Iberia/Spain I miss the definition and would propose the following hypothetical groups (very detailed and to combine, could be simplified if not distinguishable by analysis):
- FC: suspected main origin in the (Northeastern) Fertile Crescent. Suspected source populations Canaanites/Israelites (majority of Jewish J2a).
- RS: suspected main origin in the Red Sea area including Southern Levant, Northwestern Arabia and Egypt territories. Suspected source populations Nomads/Hebrews (majority of Jewish J1 and probably E).
- ME: suspected main origin in the greater Middle East area including Anatolia, Caucasus and Central Asia. Not attributable to a subarea (majority of Jewish G, T, Q, R, some others).
- Med: spread to or converted in Mediterranean lands (Central-East-Med, Iberia excluded) not later that at the Crusades. Suspected contributing source populations Greek-Roman-Byzantine (E1b-V13, J2b, ?).
- Iberic: spread to or converted in Iberian lands before or at the time of Islamic Iberia (Sephardic culture center). Speculative contributing source populations Phoenician-Celto-Iberic?
- Yiddish: spread to or converted in Northwestern France and Rhineland (Frankish homeland, Yiddish culture founders).
- E-Europe converted in Eastern Europe with focus on the Polish-Lithuanian area.
- I’m no expert of Jewish G, T, Q, R subgroups; if distinguishable a Central Asian group could be created to distinguish from Middle East (better analysis of the Khazar hypothesis)
-
If i draw conclusions in historic terms, i agree i should use more consequent names. I will have to look again to think which terms are best used. The grouping of the areas in historic terms of FC, RS, ME is probably hard. It might be possible, but a real statistical analysis is probably too time intensive for me. Side-subject: For the definition of the use of colors and originating countries i specified it in the Data-page (your point 7). I noticed just now that Armenia is reported as ME and Turkey is not reported. Since Armenia has also 3 or less persons in the dataset i should also remove it from ME.
For the analysis maybe it is best to focus on geographical distributions (of adjacent groups) without the use of “historically influenced” names. I also agree that RS and maybe also FC should be omitted and only ME used, because no clear assignation is possible (The whole area was a melting pot for over 4,000 years probably. If a group shows a probable geographical origin this could be described in the analysis text.
-
-
Many SNP markers are used to describe a group.
I think many Y-Haplogroup Admins and DTC(direct-to-consumer)-Researchers would be interested in collaborating by providing the latest terminal SNP evidence from NG-Y-sequencing results (BigY, FGC, etc.) defining Jewish sub-clusters who in the current analysis share terminal SNP/s with clusters without modern Jewish connections. I myself are interested to do so for J2 groups.
-
I thought about contacting the Admins, but have not yet done it. The J1 data is well accessible by Victar Mas, the G data is well accessible by Ray Banks. It would be nice if you will assist with the J2 groups.
-
-
A branch is recognized as an Ashkenazi branch if at least two members with different surnames are reported Ashkenazi. The majority of the group must be reported as Ashkenazi.
I would consider additional diversity criteria, likely the current ISOGG diversity criteria (STR, SNPs) and maybe tag younger groups that don’t comply with a term like “candidate” or “private” to clearly show they could be too small for general population genetics analysis. If feasible I would introduce also groups that have at least one Askenazi and one member of another Jewish group, maybe even groups with non-Ashkenazi Jews should be considered for better comparative analysis.
-
I have a few Ashkenazi Jews that are single in a group, for which i am quite sure they are Ashkenazim. In most ftdna Jewish project, it is hard for me to judge what the criteria are that a person is added on a Jewish page. In case it is: “possible Jewish ancestors”, it is too uncertain to add. Maybe i should contact the admin’s to check.
I agree that contacting Jewish-Y admins/researchers could help to improve the analysis.
-
-
The data are collected using semargl.me. This site collects data of people from ftdna.com, ysearch.com in a systematic way. In some cases people are removed from the database on request. The removal will not depend on a value for STR or on a specific branch, it is not expected to a have systematic effects. Due to the use of ftdna.com the percentage will be larger in some countries than others, e.g. larger in the United Kingdom than France. However, it is expected that it will influence all branches the same.
I agree to the selection of data collection and partly to the randomness of the data: it could be that very active genetic genealogists have motivated much more men of one group (trough coinciding surnames and places of origin) to do a Y-DNA test, seriously influencing the proportions of the groups.
-
Yes, this is possible. I could check on the maximum effect of it.
-
-
- Eastern Europe: “Ukraine”,”Poland”,”Russia”,”Lithuania”,”Latvia”,”Hungary”,”Romania”,”Belarus”
- Germanic countries: “Germany”, “Netherlands”, “Austria”, “Czech Republic”
- Southern Europe: “Spain”,”Italy”,”Portugal”,”Mexico”,”Puerto Rico”
I’m no expert of the “possible elements in the history of the Ashkenazi Jews”, but I guess it is hard to combine origins, because of the “chaotic” European migrations that occurred in the Middle Ages. Maybe Luxembourg , Belgium and France could be integrated to “Franco-Germanic“. Austria>Czechia/Slovakia>Hungary look to me like a bridge from Germanic to Eastern Europe. I would exclude Italy from “Southern Europe” and name the origin “Iberic” as Italy is probably an interesting bridge of Jewish migrations and could be reported like “Other countries”; Southern Europe suggests countries like the Balkans (Southeast Europe) or at least Greece is included.
-
for Austria-Czech-Slovakia-Hungary i had to make a choice. Reading on the historic times of these areas, and realizing the Austria and Czech have a history (after the year 1000) to be close connected to Germany. For Slovakia and Hungary the case is not so clear. Looking at all the data (see the graph in the discussion of “van Straten”) , i think there is not much difference. I could make the graphs with different countryselections, to see if it makes a difference. (I expect not; all present plots to look for these differences, did not find them; also a recent check in the jewsoffrankfurt.com project suggest the same. They have genealogy starting 15th century. The mixture between the different Y-DNA haplotypes in the different countries had already taken place). The borders between areas are difficult to draw. I thought of taking out Italy and adding Greece, but did not want to increase the amount of colors. They have 8 and 7 persons, so i could also report them separately.
Wim has the experience by his analysis. If analysis of one group using different parameters could help to check the performance I would consider it for an update.
-
The choices above [countries of origin] where checked below to see if that choice is consistent with the observed data.
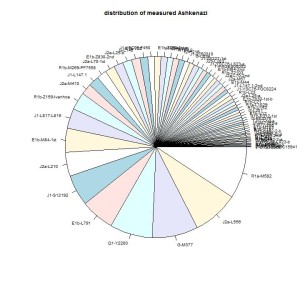
clustersincountries.jpeg gives an indication, but it would be interesting to have diagrams showing the countries of origin for every (major) branch. I have not checked the raw-data for this information and therefore could not (yet) reproduce or follow the conclusions about the “most likely Origin”
-
This is shown in the “van Straten”-discussion; it does not yet have a list of haplotypes with the numbers in the graph.
I did not study the van Straten page before. The good arguments for conclusions are interesting. In the diagram of Ashkenazim branches by size I can’t see the blue color for the Germanic countries (beside orange I see red, green and black). A color legend of the chart including the group descriptions would be both interesting and helpful.
-
-
McGee Y-Utility tool … using the mutation rates of M. Heinila (2013).
I myself for FTDNA STR-data also use mainly Y-Utility and the M. Heinila mutation rates (my Y-Utility setup files, the mutation rates used in the April 2013 publication are from April 2012 AFAIK: weightedPairs20120430.csv).
-
Recently it was found and confirmed that the amount of SNP mutations between father and son are linear dependent on the age of the father (Iceland 2012 and research). This indicates that the amount of mutations goes linear with time. It means that two generations of 25 years give the same number of mutations as one generation of 50 years. For our analysis the Heinila mutation rates should be used in combination with the generation length of 30 years. In case the average generation length of the people that were analyzed have a longer or shorter average generation length, it does not influence the calculations.
I’m not sure if the whole genome SNP mutation rate is the same or near the Y-DNA SNP mutation rate. I remind discussions that strongly debated this, as the haploid Y-chromosome undergoes much more selection (wiping out of mutations in genes), then the other recombining and diploid chromosomes; see for example Wilson-Sayres 2013 (arXiv preprint open access) compares estimates of male mutation bias using different chromosome comparisons (X/Autosome, Y/Autosome, X/Y) and observed diversity on the entire Y chromosome (1/10 of what is expected due to the effects of selection acting to reduce diversity). I have even more doubts (remind no source claiming that) if the Y-STR mutation rate behaves like SNP mutation rates or (autosomal) STR mutations rates. Would be interesting to find some sources to validate or improve this assumption about “Y-STR mutation linearity”. Maybe in the future Y-SNP mutation rate comparisons from NGS data give better predictions. 30 years as generation length seems reasonable for the average age/size of the branches (ca. 20-40 gen.).
-
Thanks, i have to read the Wilson-Sayres article.
-
-
distribution of 4 countries in comparison with the percentages of the Ashkenazi Jews.
Turkey, Armenia, Iraq and Saudi Arabia impressively show that the Ashkenazi dataset fits well in the middle. Probably they are too small for this type of diagram, but the distinction of the E1b-V13 and J2b branches would be interesting. The same distinction would be interesting for the table “Number of branches and people in haplogroups”.
-
I made a diagram to compare it with many countries, and also to compare each country with all others and determine their closest country. Many are -as expected-. Some i had not realized before. Others are difficult to determine because of the large mix in a country. I don’t see understand what you mean with “the distinction of the E1b-V13 and J2b branches would be interesting”. I have only one J2b group.
I meant E1b-V13 and J2b (because of the suspected Med/Balkanic origin) should not summarized together with the generally Middle-Eastern E1b and J2a groups.
-
-
As a measure of the amount of Jews in recent times we used the values of 700.000 in 1620 (see Demographic history of Poland), 1.000.000 in 1770 (see History of Jews in Poland) and 9.000.000 in 1890 (see Historical Jewish population comparisons)
It seems that there is no consensus about Jewish population size estimates before the end of the nineteenth century: yivoencyclopedia.org for Polish–Lithuanian Commonwealth in thousands: 1495: 10, 1550: 55, 1700: 285, 1765: 750 (Sources: adapted from Mark Kupovetsky, 1983, pp. 75–76; 1985, p. 75; and 2006; Laszlo, 1966, pp. 89–90; Stampfer, 1997 and 2003; Weinryb, 1973, pp. 309–320; and the author’s additional estimates.).
-
You are correct. I took numbers that seemed most reliable to me. The quality before 1620 is very bad. After that i read “van Straten” a month ago, i realized that the quality of the years 1620 and 1770 is probably also worse than i expected. I don’t think it influences the conclusions, but it is good to have a look again, take the best values, and indicate how sensitive the conclusions depend on these values.
The difference of the data seems not big, but a check how much this influences conclusions is interesting.
-
-
grow rate Models
I do not feel qualified to comment on this currently. It looks like consistent analysis interesting to reproduce on different datasets.
-
🙂
-
-
Ashkenazi: the relation with other branches
In this page we will describe what we know about the relation of the Ashkenazi Y-DNA branches and what is likely the relation to other branches.
This looks like a very interesting analysis, but I had difficulty both to access the images (sizeofthe40branchesandbeforebig.jpeg, orderbysize.jpeg) with Firefox and to extract quickly useful information out of them. Maybe I really need to go trough the raw-data at least for J2.
-
maybe is should use a tool so people can easier zoom in and out of the data. Does figshare give me this option?
The underlying raw-data (CSV?) could be helpful. For PDF-images figshare allows to Enlarge – I expect the same feature for other image formats.
-
-
The largest groups are in order or size: R1a-M582, J2a-L556, G-M377, Q1-Y2200, E1b-L791. These five groups are about 35-40% of the Ashkenazi Jews. These groups have characteristics that are different from the groups that passed Iberia. The differences are: the size of the group is larger, the STR-variablity is larger, there are no nearby branches leading to other locations (with the possible exception of Q1-Y2200).
The characteristics suggest that they came early, and they did not pass Iberia in the period of the Caliphate of Cordoba. The other groups that passed Iberia have many converso descendants in Iberia; these five groups don’t have that. During the period of the Caliphate there was probably no reason to leave the Caliphate and settle elsewhere. Only one of the five groups (Q1-Y2200) has several relative close descendants in other areas (Kazakhstan, Turkey, Iberia, Britain, Middle East, Algeria). The other four groups have a narrow line in the phylogenetic tree before the expansion of the groups started. One reason can be that the ancestors of the four groups lived in an area where the population did not grow.
If the Y-DNA ancestors of these groups came from Khazaria, it remains open if they came in Khazaria from Judea or their ancestors were Khazars and they became Jewish in the Kingdom of Khazaria. With the limited data available, it is also possible they came from another area to the Polish Lithuanian area.This is one of the most interesting Questions. Extensive comparison with Mizrahi and other non-European Jewish Y-DNA data would be interesting, probably there is not enough public data.
-
I completely agree. I think there must interesting Mizrahi and non-European Jewish Y-DNA data, but i don’t know where to find it. Maybe it is not available. There is no ftdna Mizrahi project.
Hopefully contacting Jewish-Y admins/researchers will clear out if data is available.
-
-
J2a-L556 J2a-M92 J2a-M67 J2a-L210 J2a-L70-1st J2a-L70-2nd J2a-L25-a J2a-L25-b J2a-M410 J2b-L283 In my J2 research I do not focus on J2a-L24 (incl. L25, L70), J2a-M67 (incl. L556, L210) and J2b branches, as there are other interested/qualified admins/researchers for those areas. An exception is the J2a-L210 group because of a Tyrolean sample which like other L210+ kits does not cluster nearly to the Jewish branch. I will address my interest in this and a likely improvement of the J2 terminal clades with a dedicated research and analysis.
-
Thanks, it would be nice to improve the J2a-L210 group.
I will also work on the other J2 groups, but my main interest will be in the wide L210 group.
-
-
Q: Carmi et al. published in August 2014 a research on the history of the Ashkenazim based on auDNA data. Are the conclusions of this article consistent with the analysis of the analysis of this website?
A: Yes, the majority of the conclusions (number of Ashkenazi ancestors, time that this group changed from population bottleneck to strong population growth and average population growth) are consistent. See the a review of the article and a comparison of the results.I think Cochran improved the Carmi et al. study estimates. Regarding the autosomal European ancestry (admixture component/s) of Ashkenazi I know of estimates from 30-50% (Atzmon et al, Behar et al), so additional studies are welcome to confirm this substantial difference from Y-DNA origins (only about 6% European according to Wim’s analysis) and this also makes questionable the FAQ answer that “auDNA and Y-DNA … will give similar results”. Ashkenazi mtDNA likely has a majority (80%) of European ancestry (Costa et al. 2013) acquired since BC in South Europe and later in Central Europe.
If this data for au/at, Y and mt genomes is valid, the following scenario might the most plausible: Jewish communities of emigrants from the Middle East established themselves in the Greek and Roman South European area. The men over the centuries married with local women and to a much smaller extent local men where assimilated into the communities. The stability and prosperity of the Frankish empire attracted many Jews from Southern Europe and those communities in the Middle Ages became the center of the Yiddish culture, still admixing with local people (mainly women, likely a few men). Stability and protection in Poland caused the immigration of many Jews from Western and Southern Europe, where prosecution, segregation and exile did became common. This migration to Eastern Europe endured for a longer period of time combined with strong population growth. Admixture contributions from local people and/or former Khazar territory did happen. Trough all the time of the “European Jews” it seems that admixture between geographically distant communities was not seldom, emphasizing the strong network between this minority.-
I have not yet read the Cochran contribution. I will have a look. The admixture of 6% European that i mentioned is 6% that is clear. For e.g. the five largest groups, it is possible they came from the Kingdom of Khazaria, and they did not come from from ME before. If you count these as European, the percentage goes up dramatically.
I have not yet read the Costa paper. I saw another reference today to the Costa paper. I will read it. Thanks.
I would be somewhat surprised by a scenario in which the maternal lines are very different from the male lines. Somehow i expect these differences in many cultures, cut i don’t expect it in the Jewish tradition. I might well have to adjust this thought of mine. 😉I think analysis of (future) data including NGS-Y-Sequences from the groups sharing the most recent Y-Ancestor with Ashkenazi/Jewish groups is essential and the same will be necessary for mt and autosomal DNA (full genomes = all variance analyzed). I think we can not hope ancient DNA finds will resolve the questions in the near future, tough anyone certainly hopes Jewish remains 500-2,500 years old will be found and analyzed.
-
-
Reference to this site: bit.ly/40Ashkenazim > dl.dropboxusercontent.com/u/3487498/…
If Wim feels comfortable with it and would like to publish to “more stable and recognized” places I strongly suggest to use figshare and/or a preprint place like bioRxiv. The same maybe is valid for the also interesting, but not studied (yet) in detail by me paper on 2000 years genetic variability in Flanders, Brabant and Limburg.
Last but not least I do miss a time indication (year): best would be to indicate the date of data collection, main analysis and writing of main conclusions.-
I would like to publish it to “more stable and recognized”. I submitted my Z640 paper to http://rjgg.molgen.org/index.php/RJGG at the start of this year. When i got it back with comments, i was analyzing the new dataset, and have not yet adjusted the Z640 paper. This website is a consequence of my previous analysis on J1-Z640.
If RJGG allows preprint and re-publication elsewhere (for example the CC-BY license does that) there should be no limit to do so. In accordance with widest possible discussion and a citable place (DOI) I think this is what citizen scientists want.
-

My brother’s Ydna haplogroup is Q1b1a ((-L245) , for David Goldfoot. Our grandfather was Nathan Abraham Goldfus and changed the name after leaving Telsiai, Lithuania. Goldfus is of German origin. I have found through familyfinder, an autosomal test to be connected to the Rabbi of Worms, Germany who in turn was connected to RASHI, a rabbi of France/Germany born 1040-died 1105. Oral history of RASHI is that he was related to King David of Israel. Our Q Ashkenazi lineage is only 5% of the Jewish population.
If it can help you in any way, I”ve created a time line on one of my blogs with people and events on it. http://israel-nadene.blogspot.com/2014/08/jews-living-in-judea-samaria-and-other.html
I’m the administrator for surname Goldfoot at FTDNA, and we have very few people, but luckily I did find a branch I didn’t know about who went from Lithuania to England and Ireland, which my grandfather also did, but then they went to South Africa while my grandfather went to the USA. We didn’t know about them at all until my genealogy searching made contact.
If your Y-lineage kit/cluster is not present in Wim Penninx’s study please contact him for a possible update.
I am j2a4h2. My oldest known paternal ancestor was from Switzerland in the 1400’s.
We have no knowledge of Jewish ancestry – as far as we know we were always Christian.
But there were many Hollingers (same name) killed in the holocaust. Given the J2a haplogroup it sound likely that our ancestors were Jewish.
More on the blog below:
http://hullingerdna.blogspot.com/
I think it is better if you always add your terminal SNP to the haplogroup name, J2a4h2 is not anymore currently used (by FTDNA). As you are J2a-F761 I see you have two Italian matches in J-L24 project. You should focus on the history of all lineages in this small cluster and try to find more and check if one of them is or was Jewish, otherwise you will remain with speculation about the own lineage history.